Ranging from understanding gene regulatory relationships better to further increasing the multiplexing capabilities of single cell genomics technologies, BBI member labs share their research through multiple publications in a variety of scientific journals. Two researchers, University of Washington graduate student in the Department of Genome Sciences, Florence Chardon and Dr. Molly Gasperini, Scientist at Octant Bio and an alumni of the University of Washington Department of Genome Sciences, reflect on four scientific publications that they feel made the biggest impact in 2019 from BBI member labs.
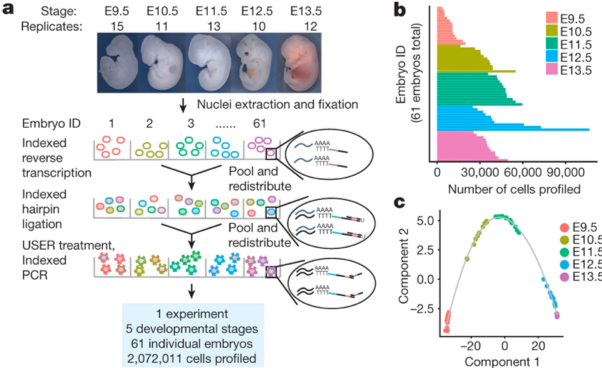
Multiplexing single cell measurements has allowed researchers to gain unprecedented insight into biology. Multiplexing allows researchers to look at not one, but many different measurements at once. The power of multiplexing is illuminated in two of our top four publications, “The single-cell transcriptional landscape of mammalian organogenesis” and “Massively multiplex chemical transcriptomics at single-cell resolution.” Both publications utilize single-cell combinatorial indexing RNA-sequencing, an RNA sequencing technology developed by Dr. Junyue Cao, to profile up to two million cells in a single experiment.
When asked what was most exciting about the mammalian organogenesis work, Chardon comments, “Sci-RNA-seq-3 allows for transcriptional profiling of unprecedented number of cells at low cost. In addition to elucidating the transcriptional landscape of mammalian organogenesis, this work unleashes a powerful new single-cell RNA sequencing technology that can be used to better understand the transcriptional consequences and responses to many developmental processes and functional perturbations.” Dr. Molly Gasperini states, “paper presents one of the first atlases of the mammalian organ development at single cell resolution, as well as provides a novel methodology.”
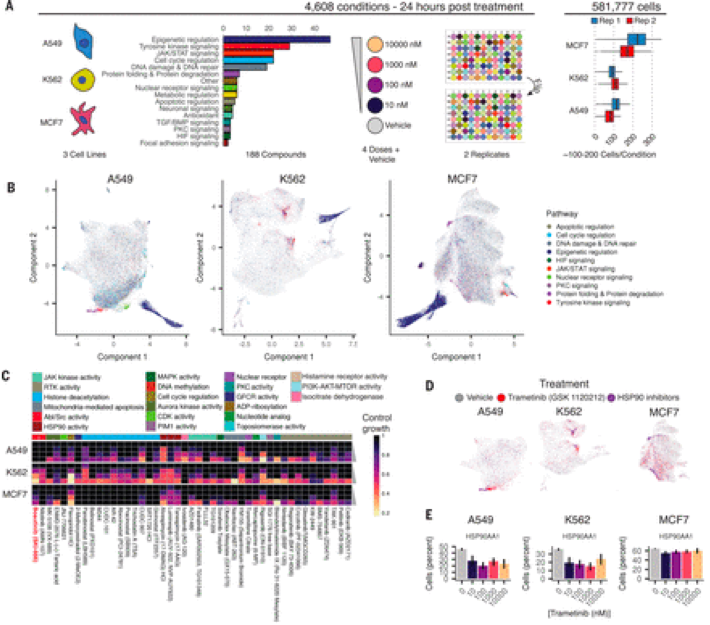
The ability of sci-RNA-seq-3 to transcriptionally profile millions of cells in a single experiment allowed the trio of Sanjay Srivatsan, José McFaline-Figueroa, and Vijay Ramani to develop sci-Plex. Chardon comments on why sci-Plex represents such a huge leap in how we understand cellular responses to drugs, “In a field where multiplexed technologies have allowed for the interrogation of large numbers of perturbations and single cells in single experiments, this work provides a dramatic advance in how we multiplex single-cell genomics experiments and use these experiments to study previously poorly understood, complex biology. Cellular responses to drug treatments have traditionally been categorized in binary ways, such as cell life or cell death, and now with sci-Plex, we can better understand the heterogenous transcriptional ways in which cells respond to drug treatments.”
 José McFaline-Figueroa is the lead author on another publication titled, “A pooled single-cell genetic screen identifies regulatory checkpoints in the continuum of the epithelial-to-mesenchymal transition.” When asked what was most impressive about this work, Gasperini comments, “This paper applies a cutting edge method (single-cell CRISPR screens) to gain new insight into a cancer and cell development.” Chardon adds, “This work demonstrates the power of pooled single-cell genetic screens to better understand the genetic circuitry that underlies certain biological processes. The biological process studied here is the epithelial to mesenchymal transition, one of critical importance in cancer development, but moreover this work introduces a framework by which to study these processes.”
The fourth publication which we chose as one of the most impactful publication of 2019 is “Saturation mutagenesis of twenty disease-associated regulatory elements at single base-pair resolution.” This work serves to address the criticality of gaining a more thorough understanding of noncoding regions of the genome, as the majority of common variants associated with disease fall in these exact regions. This work uses a massively parallel reporter assay framework to test the effects of 30,000 single nucleotide substitutions and deletions in promoter and enhancer regulatory regions. This work provides a significant advance in cataloguing noncoding regulatory elements by measuring the functional effects resulting from mutations in these regions.
José McFaline-Figueroa is the lead author on another publication titled, “A pooled single-cell genetic screen identifies regulatory checkpoints in the continuum of the epithelial-to-mesenchymal transition.” When asked what was most impressive about this work, Gasperini comments, “This paper applies a cutting edge method (single-cell CRISPR screens) to gain new insight into a cancer and cell development.” Chardon adds, “This work demonstrates the power of pooled single-cell genetic screens to better understand the genetic circuitry that underlies certain biological processes. The biological process studied here is the epithelial to mesenchymal transition, one of critical importance in cancer development, but moreover this work introduces a framework by which to study these processes.”
The fourth publication which we chose as one of the most impactful publication of 2019 is “Saturation mutagenesis of twenty disease-associated regulatory elements at single base-pair resolution.” This work serves to address the criticality of gaining a more thorough understanding of noncoding regions of the genome, as the majority of common variants associated with disease fall in these exact regions. This work uses a massively parallel reporter assay framework to test the effects of 30,000 single nucleotide substitutions and deletions in promoter and enhancer regulatory regions. This work provides a significant advance in cataloguing noncoding regulatory elements by measuring the functional effects resulting from mutations in these regions.


