
Data processing pipeline
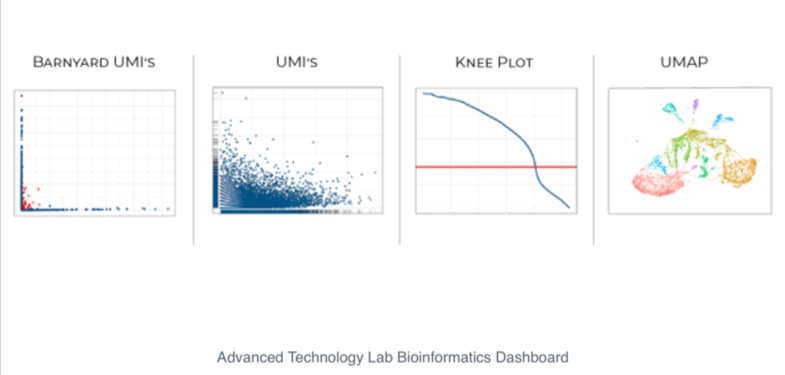
The Brotman Baty bioinformatics team preprocesses all single cell sequencing data and returns data ready to use, including a web dashboard that shows the results of some basic quality control and analysis. We return all raw data as well as a premade Cell Data Set (cds) object to use with Monocle3 , a well-developed toolkit for analyzing single-cell gene expression experiments. Basic Monocle3 analyses and quality control steps are outlined in the web dashboard, allowing investigators with limited R programming experience to jump right into data analysis.
Learn more about the method, tools, and BBI services here>
For existing clients of SCI services, view your project status here >
Single Cell Training & Troubleshooting
Training Session December 2021
[BBI Tutorial Resources](https://github.com/bbi-lab/bbi-training-resources) This repository stores code used in the quarterly BBI single-cell data analysis tutorials.Training Session February 2021
BBI Tutorial Resources This repository stores code used in the quarterly BBI single-cell data analysis tutorials.
Training Session December 2020
BBI Tutorial Resources This repository stores code used in the quarterly BBI single-cell data analysis tutorials.
Training Session September 2020
BBI Tutorial Resources This repository stores code used in the quarterly BBI single-cell data analysis tutorials.